Finally Automate
Your Business
Finally Automate
Your Business
Unlock the power of integration
Unlock the power of integration
Trusted by Others
Trusted by million+ users



Discover the Benefits
Seamless Integration,
Unparalleled Efficiency

Seamlessly add contacts from GHL to your membership platform and vise versa.

Automatically collect 5 star Google reviews from your members.

Communicate with your members and prospects on autopilot.

Manage your business more efficiently & make data-driven decisions.
WHY YOU NEED THIS
Before vs After SYNX
Manually transferring contacts.
Manually tagging new members.
Manually adding leads to follow ups.
Human error & forgetting about leads.
Manually sending review requests.
2 way book classes / appointments.
Automated google review requests.
2 way contact creation.
Automated membership signups.
Fully pre built snapshot for GHL.

Capture Leads
Discover the ease of lead capture with SYNX. Say goodbye to the hassle of 10-step signups and hello to effortless customer acquisition.

A Better Experience
Elevate customer journeys with deep insights and effortless communication.

A Better Experience
Elevate customer journeys with deep insights and effortless communication.

Track Your LTV
Seamlessly track your customers spend & know how much your customers are worth.

Simplified Automation
Revolutionize your business operations! Trigger automations based on customer behaviors and watch your retention soar.

Data Management
Say goodbye to the hassle of juggling various platforms and the headache of aligning mismatched data sets.

Data Management
Say goodbye to the hassle of juggling various platforms and the headache of aligning mismatched data sets.

Attract New Customers
Effortlessly empower your customers to become your most influential advocates, magnetizing new clientele.
Fully Automated Class Bookings

Automated Class
Reminders For Newbies
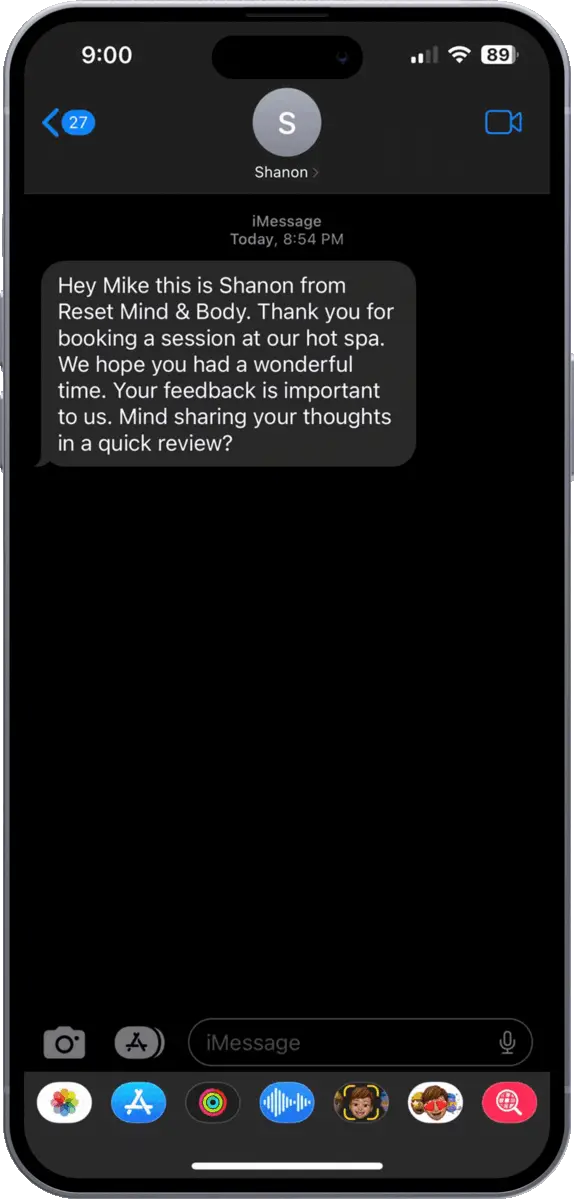
Bridge the gap, connect with new leads, and empower them to effortlessly book their first class with you. Say goodbye to pre-class jitters as you guide them towards an exciting and unforgettable experience.

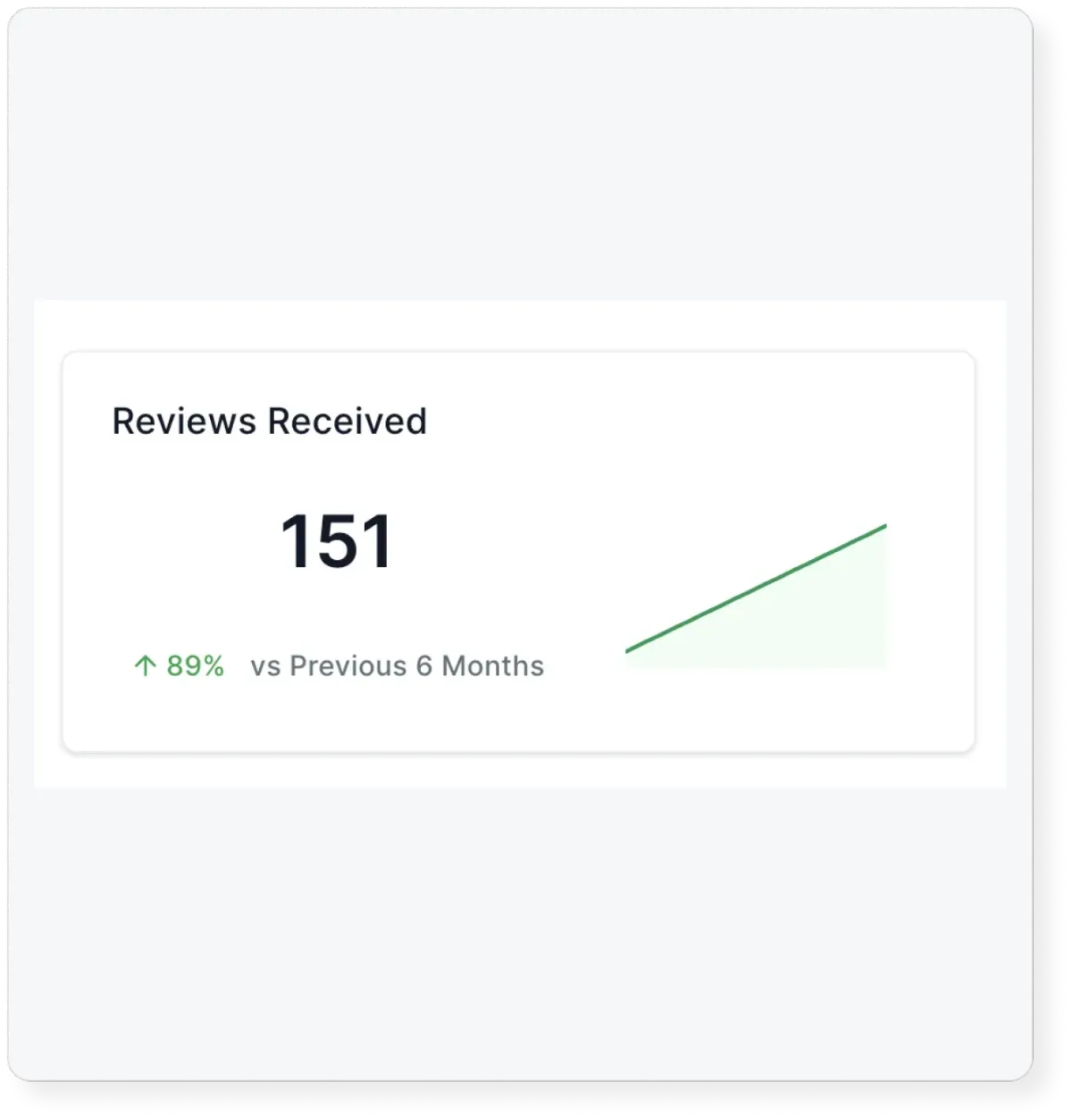

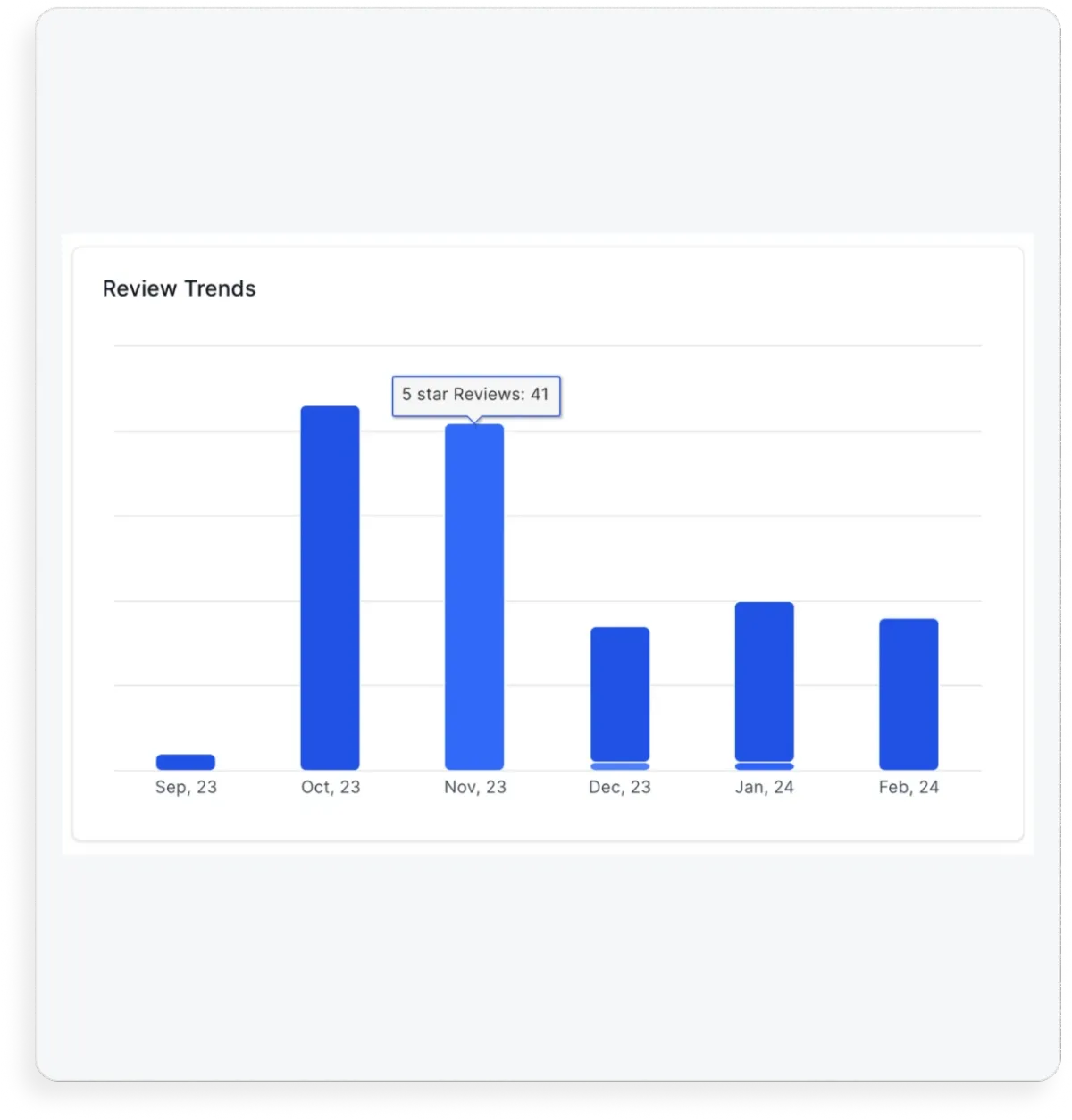
Collect Reviews On Autopilot

Bridge the gap, connect with new leads, and empower them to effortlessly book their first class with you. Say goodbye to pre-class jitters as you guide them towards an exciting and unforgettable experience.

Why Your Old Reviews
Are Irrelevant

95% of new customers read online reviews before buying and 85% of those consumers don’t think older reviews are relevant. Studies suggest that 80% of your new online reviews are going to originate from follow-up with them. SYNX makes this effortless.
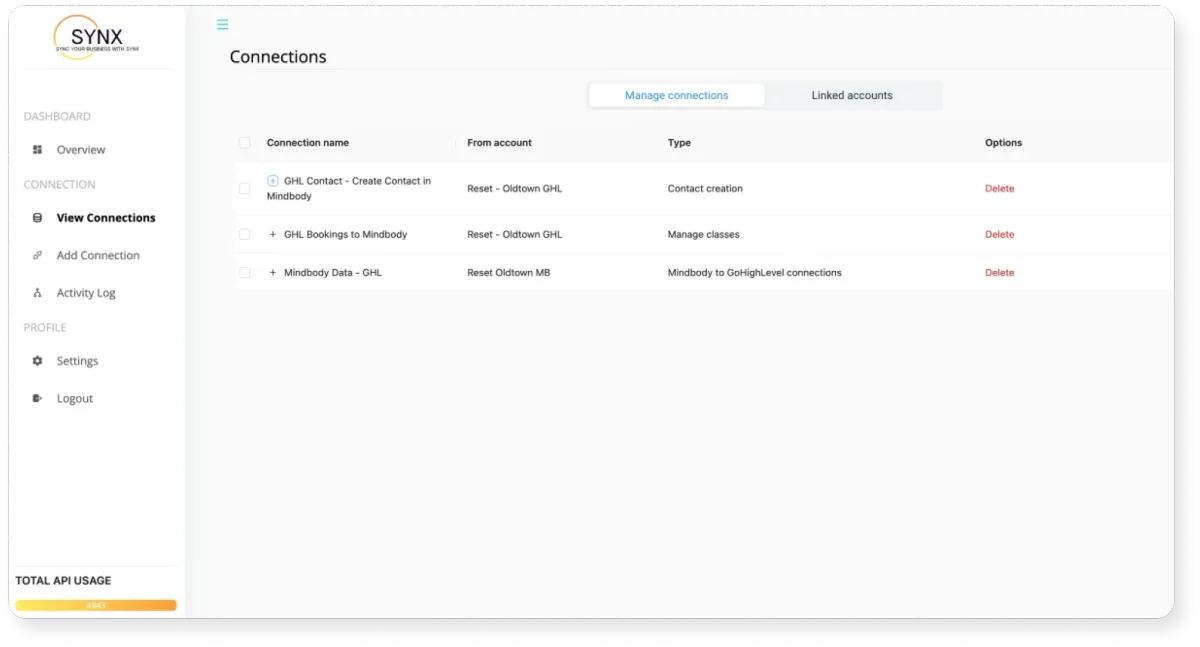
We Make It Easy
To Automate.
Plug & play TEMPLATE

Harness the full potential of both systems and optimize your operations. See for yourself why other business owners are raving about how SYNX helps them scale their revenue while spending less time working on mundane tasks.
PRICING
Choose Your Plan
Monthly
2 way class / appointment booking & cancelations
Track signed in / no shows
2 way contact creation
Membership signups
Membership cancelations
Pre built snapshot for you GHL account
Today just

Yearly
2 way class / appointment booking & cancelations
Track signed in / no shows
2 way contact creation
Membership signups
Membership cancelations
Pre built snapshot for you GHL account
Today just

Our Integration Partners
SYNX directly integrates with you GHL account and your membership
and booking platforms to collect the actions your clients are taking.
Don't see an integration your looking for? Let Us Know Whats Missing!

Coming Soon
WHY YOU NEED THIS
The Ultimate Solution For Seamlessly Syncing Data With GoHighLevel.
STILL NOT SURE?
Our Satisfaction Guarantee
Harness the full potential of both systems and optimize your operations. See for yourself why other business owners are raving about how SYNX helps them scale their revenue while spending less time working on mundane tasks.

Frequently Asked
Questions
We'll help you save countless hours on manual data entry and aggregate all the data you need to focus on closing deals quickly and seamlessly
Who can benefit from using SYNX CRM?
Companies who use membership management platforms like Mindbody, Mariana Tek, and Club Ready and also use Go High Level as their CRM and want to be able to connect the two platforms together for a 2 way sync.
What are the different pricing plans for SYNX CRM?
You can get a SYNX account for $50/Month. If you are a franchise looking for bulk pricing, please reach out to our sales team.
Is there a free trial available, and what does it include?
Yes, there is a 14 day free trial and it includes full account access with zero restrictions!
What kind of support can I expect if I encounter an issue?
You can reach out to our support team or visit our help center to trouble shoot the issue. You can expect us to make sure we figure out what the issue is and to get it fixed.
Free 14 Day Trial
Create your free account and try it for yourself to see how you like it!

